USP19 Antibody (C-term) Blocking Peptide
Synthetic peptide
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND
| Primary Accession | O94966 |
|---|---|
| Other Accession | NP_006668 |
| Clone Names | 3072510 |
| Gene ID | 10869 |
|---|---|
| Other Names | Ubiquitin carboxyl-terminal hydrolase 19, Deubiquitinating enzyme 19, Ubiquitin thioesterase 19, Ubiquitin-specific-processing protease 19, Zinc finger MYND domain-containing protein 9, USP19, KIAA0891, ZMYND9 |
| Target/Specificity | The synthetic peptide sequence used to generate the antibody AP2145b was selected from the C-term region of human USP19 . A 10 to 100 fold molar excess to antibody is recommended. Precise conditions should be optimized for a particular assay. |
| Format | Peptides are lyophilized in a solid powder format. Peptides can be reconstituted in solution using the appropriate buffer as needed. |
| Storage | Maintain refrigerated at 2-8°C for up to 6 months. For long term storage store at -20°C. |
| Precautions | This product is for research use only. Not for use in diagnostic or therapeutic procedures. |
| Name | USP19 |
|---|---|
| Synonyms | KIAA0891, ZMYND9 |
| Function | Deubiquitinating enzyme that regulates the degradation of various proteins by removing ubiquitin moieties, thereby preventing their proteasomal degradation. Stabilizes RNF123, which promotes CDKN1B degradation and contributes to cell proliferation (By similarity). Decreases the levels of ubiquitinated proteins during skeletal muscle formation and acts to repress myogenesis. Modulates transcription of major myofibrillar proteins. Also involved in turnover of endoplasmic- reticulum-associated degradation (ERAD) substrates (PubMed:19465887, PubMed:24356957). Mechanistically, deubiquitinates and thereby stabilizes several E3 ligases involved in the ERAD pathway including SYVN1 or MARCHF6 (PubMed:24356957). Regulates the stability of other E3 ligases including BIRC2/c-IAP1 and BIRC3/c-IAP2 by preventing their ubiquitination (PubMed:21849505). Required for cells to mount an appropriate response to hypoxia by rescuing HIF1A from degradation in a non-catalytic manner and by mediating the deubiquitination of FUNDC1 (PubMed:22128162, PubMed:33978709). Attenuates mitochondrial damage and ferroptosis by targeting and stabilizing NADPH oxidase 4/NOX4 (PubMed:38943386). Negatively regulates TNF-alpha- and IL-1beta- triggered NF-kappa-B activation by hydrolyzing 'Lys-27'- and 'Lys-63'- linked polyubiquitin chains from MAP3K7 (PubMed:31127032). Modulates also the protein level and aggregation of polyQ-expanded huntingtin/HTT through HSP90AA1 (PubMed:33094816). |
| Cellular Location | Endoplasmic reticulum membrane; Single-pass membrane protein. Note=Accumulates in the mitochondria-associated ER membrane (MAM) in response to hypoxia |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
Background
Modification of target proteins by ubiquitin participates in a wide array of biological functions. Proteins destined for degradation or processing via the 26 S proteasome are coupled to multiple copies of ubiquitin. However, attachment of ubiquitin or ubiquitin-related molecules may also result in changes in subcellular distribution or modification of protein activity. An additional level of ubiquitin regulation, deubiquitination, is catalyzed by proteases called deubiquitinating enzymes, which fall into four distinct families. Ubiquitin C-terminal hydrolases, ubiquitin-specific processing proteases (USPs),1 OTU-domain ubiquitin-aldehyde-binding proteins, and Jab1/Pad1/MPN-domain-containing metallo-enzymes. Among these four families, USPs represent the most widespread and represented deubiquitinating enzymes across evolution. USPs tend to release ubiquitin from a conjugated protein. They display similar catalytic domains containing conserved Cys and His boxes but divergent N-terminal and occasionally C-terminal extensions, which are thought to function in substrate recognition, subcellular localization, and protein-protein interactions.
References
Nagase, T., et al., DNA Res. 5(6):355-364 (1998).
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.




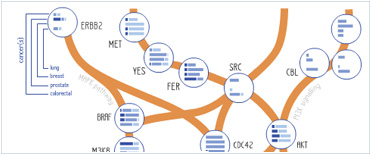







 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.